Pretreatment Procedure for metabolomics (Biological sample)
Pretreatment Procedure for the Blood Serum
Here you will find an overview of pretreatment procedures for determining primary metabolites in serum using GC-MS or LC-MS. Please refer to the "Pretreatment Procedure Handbook for Metabolites Analysis" for details on the reagents and experimental equipment used.
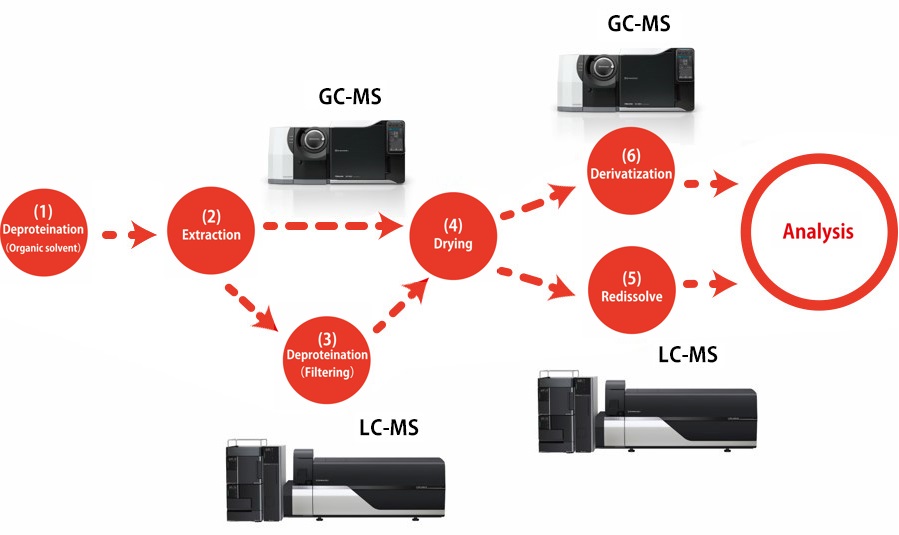
This method*1 is used to extract mostly hydrophilic metabolites by adding an extraction solvent consisting of a mixture of water, methanol, and chloroform to deproteinize the liquid sample and then recover the water layer. Because only vaporized compounds are detected by GC-MS analysis, compounds that do not vaporize easily must be derivatized. Derivatization is performed in two stages by methoximation and trimethylsilylation (TMS).
(1) Deproteination with organic solvent
Collect a 50 µL sample of blood serum in a 1.5 mL tube (Fig. 1). If an internal standard is used, add the internal standard to the tube. Add a certain amount of solvent mixture containing water, methanol, and chloroform at a ratio of 1:2.5:1 (extraction solvent) to the tube. When the protein denatures, the tube contents turn a cloudy white color (Fig. 2). It is convenient that the extraction solvent can be prepared in large quantities in advance and stocked in 1-L reagent bottles, or other containers, at ambient. The extraction solvent can also be added to the tube after premixing it with the internal standard solution in a 50 mL tube. After mixing thoroughly with a vortex mixer (Fig. 3), heat the mixture to 37 degree Celsius and shake it for 30 minutes at about 1,200 rpm in a heated shaker (Fig. 4). When finished shaking, centrifuge the mixture for three minutes at 4 degree Celsius and 16,000 G (Fig. 5). The solution is separated into two layers, with the denatured protein precipitated to the boundary surface between the layers (Fig. 6). Obtain 225 µL of the supernatant by carefully inserting a pipette tip into the tube, so that the tip does not contact the precipitate or the chloroform layer, and place it in a new tube (Fig. 7).
(2) Extraction of hydrophilic metabolites
Add a certain amount of ultrapure water to the new tube containing the recovered supernatant. Proteins still remaining in the solution are denatured, turning the solution a cloudy white color (Fig. 8). After mixing thoroughly in a vortex mixer (Fig. 9), centrifuge the mixture again for three minutes at 4 degree Celsius and 16,000 G. After centrifuging (Fig. 10), obtain the supernatant and place it in a new tube (Fig. 11).
(3) Deproteination using size exclusion filter (Only the case of LCMS)
In LC-MS analysis, the organic solvent is applied as the mobile phase, exposing the sample to a concentrated organic solvent environment during the analysis. If proteins that were not fully deproteinized precipitate during analysis, it could cause line blockage that stops the analysis. To avoid that situation, size exclusion filtration is used after extraction to eliminate proteins. To remove the denatured proteins between the layers, attach the size exclusion filter to a clean tube (Fig. 12), add a certain amount of the supernatant to the top of the filter and close the cap. Centrifuge 60 minutes at 4 degree Celsius and 16,000 G (Fig. 13). After centrifuging, remove the filter from the tube.
(4) Drying
Poke two or three small holes in the cap of the 1.5 mL tube and cut the cap from the tube (Fig. 14). Attach the cap to the tube containing the collected supernatant (Fig. 15). Use the centrifugal evaporator to evaporate the methanol from the solution for 25 minutes (Fig. 16). After evaporating for 25 minutes, place the tube in the deep freezer without changing the cap. Let it sit for about 15 minutes and then confirm that the solution is fully frozen. Finally, dry it in the freeze dryer (Fig. 17). If it is necessary to store the sample after pretreatment, store the dried sample in the deep freezer for the LC-MS analysis sample, and store it in the room temperature desiccator for the GC-MS analysis sample. This is because derivatization is necessary for analysis by GC-MS, and desorption efficiency decreases when moisture is absorbed.

(5) Redissolve (Only the case of LCMS)
(6) Derivatization (Only the case of GCMS)
Weigh the methoxyamine hydrochloride. Dissolve the weighed methoxyamine hydrochloride in pyridine to make a concentration of 20 mg/mL (Fig. 20). Prepare enough methoxyamine-pyridine solution for the given number of samples involved, assuming 80 µL is used per tube.
* The methoxyamine hydrochloride may be difficult to dissolve in some cases. If undissolved residue is visible, use a sonicator or other means to ensure it is completely dissolved.
A solid substance with a white to whitish-yellow color will be clinging to the sample tube walls after freeze drying (Fig. 21). Add 80 µL of the 20 mg/mL methoxyamine-pyridine solution (Fig. 22) and mix it in the sonicator until the residue is dispersed (Fig. 23).
* Any moisture contained in the sample will decrease derivatization efficiency, so be especially careful to prevent water from entering the sample
Heat and shake the sample in the heated shaker for 90 minutes at 30 degree Celsius and about 1,200 rpm (Fig. 24). Then add 40 µL of MSTFA (Fig. 25) and heat and shake the sample in the heated shaker for 30 additional minutes at 37 degree Celsius and about 1,200 rpm. If any residue remains, centrifuge at 16,000 G for three minutes (Fig. 26), collect the supernatant in a GC-MS vial, and use it for analysis (Fig. 27).