Detection of Mold and Yeast Genes
Mold and yeast exist throughout the environment and there are many beneficial varieties of each in food sources such as miso (fermented soybean paste), cheese, bread, wine, and sake, etc., and those that are mildly to extremely toxic.
Polymerase Chain Reaction (PCR) technology has been used in recent years as one of the molecular biological techniques for detecting fungi such as molds and yeasts.
When PCR is performed, a complicated pretreatment operation is typically required to extract the DNA from fungus samples like molds and yeasts. Here we introduce an example of direct mold and yeast analysis using the MCE-202 "MultiNA" DNA/RNA analyzer based on microchip electrophoresis technology, in which the PCR reagent "Ampdirect® Plus" is used to enable the PCR reaction without the need for any PCR pre-processing operations.
Analytical Procedure
Samples consisted of two types of mold and one type of yeast, as shown in Table 1.
The genusEurotiumis a type of mold which grows in dried foods, such as dried goods, bread, filled buns, and jam, etc. The genusPenicillium, also referred to as "blue mold", is a genus of mold that occurs in many types of food, such as citrus fruits, grains, and dairy products. There are various types ofPenicillium, ranging from beneficial varieties that are used in foods such as in the production of cheese, to harmful types such as toxic mold.Saccharomyces cerevisiaeis budding yeast, and includes such varieties as baker's, wine, and sake yeast.
For the PCR reaction reagent, we used Shimadzu's "Ampdirect® Plus Enzyme Set" for gene amplification, and the PCR reaction conditions used are listed in the included instruction manual.
The mold and yeast that had been cultured in agar medium were attached to a micropipette tip, then suspended in the PCR reagent solution, and PCR was conducted (Fig. 1).
Primers (ITS primers, for fungi, designed for quick identification of microorganisms by genetic analysis as described in the Japanese Pharmacopoeia(2)) were used for detection of ITS regions(1) when performing PCR.
The PCR products were analyzed using the MultiNA. (Fig. 2)
|
(1) |
The Internal Transcribed Spacer (ITS) regions refer to the 2 regions between the 3 ribosomal RNA genes (rDNA), 18 S, 5.8 S and 28 S, (ITS 1 between 18 S and 5.8 S, ITS 2 between 5.8 S and 28 S). It is known that there are differences in the base sequences in these ITS regions among different kinds of bacteria. |
|
(2) |
The Japanese Pharmacopoeia is book containing standard criteria for pharmaceutical products intended to provide guidelines for establishing standard properties and quality of pharmaceutical products. |
Reagents/Kits
- Ampdirect® Plus Enzyme Set
- DNA-500 Kit
- SYBR® Gold nucleic acid gel stain
- 25bp DNA Ladder
Analytical Conditions for PCR Products
|
Analytical Instrument |
: MCE-202 "MultiNA" |
|
Analysis Mode |
: DNA-500 Pre-mix mode |
The analysis results for the mold and yeast gene ITS regions obtained according to the procedures of Fig. 1 and 2 are shown in Fig. 3.
The amplification products specific to the genes of the respective molds and yeast were clearly detected (The size estimations shown in the electropherogram in this figure were obtained through actual experimentation). The measurement results using the MultiNA were obtained as a gel image and electropherogram.
In addition, the estimated size values for the amplification products were calculated using a calibration curve generated using a standard sample (Ladder) with known sizes, and the estimated concentration values were calculated based on a high molecular internal standard marker (Upper Marker) of known concentration used as a standard. Thus, evaluation of the target amplification products are detected more reliably and easier than when using agarose gel electrophoresis.
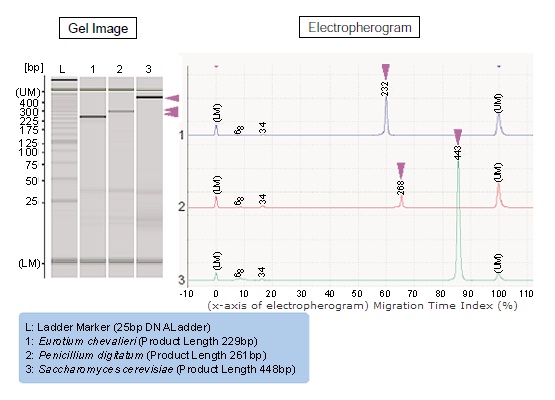
Fig. 3 Analytical Results for Mold and Yeast Genes
Microchip Electrophoresis System for DNA/RNA

DNA and RNA samples are separated by size by the electrophoresis system using a microchip so that the size of nucleic acid (DNA/RNA) samples is verified and approximately quantitated. The microchip achieves an electrophoresis system capable of speedily conducting electrophoresis separation, and a fluorescent detector ensures that analysis is performed to high sensitivity and, moreover, fully automatically.


